9. More useful functions
This section introduces many amazing functions to make the data analysis workflow streamlined.
9.1 str_detect_multi
The function str_detect_multi could help multi-detect the presence/absence of a match.
①The function str_detect_multi can match multiple strings of interest at the same time.
②The parameter
exact support match partly and completely.text <- c('Bacilli_unclassfiled','Bacteroidia_uncuture','Other')
str_detect_multi(text,c('Bacilli','bacteroidia'),exact=FALSE) # Ignore the capital letter
str_detect_multi(text,c('Bacilli','Bacteroidia'),exact=TRUE) # Set the matched completely
In addition, str_detect_multi can also help in the EasyMultiProfiler data analysis pipeline.
🏷️Example 1:Extract microbial data from the Bacteroidetes and Firmicutes at Phylum level.
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_filter(feature_condition = str_detect_multi(Phylum,c('Bacteroidetes','Firmicutes')))
🏷️Example 2:Kick out the taxa without annotation.
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_filter(feature_condition = !str_detect_multi(Class,'unclassified'))
9.2 EMP_to_EMP1
In the EasyMultiProfiler data analysis process, the function could easily export microbial data into a data list for the EasyMicroPlot package.
①When exporting data, it is necessary to add full annotations in EMP_feature_convert.
②The function could not work before collapsing microbial level.
🏷️Example:Export microbial data and perform co-occurrence network analysis in the EasyMicroPlot package.
# Get the data from EasyMultiProfiler
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_feature_convert(from = 'tax_single',add = 'tax_full') |>
EMP_to_EMP1(estimate_group = 'Group') -> deposit
# Work in the EasyMicroPlot
library(EasyMicroPlot)
cooc_re <- cooc_plot(data = deposit$data,design = deposit$mapping,
meta = deposit$meta,min_relative = 0.001,
min_ratio = 0.7,cooc_method = 'spearman',
cooc_output = T)
9.3 top_detect
This function helps the EasyMultiProfiler data analysis pipeline quickly filter out the desired highest or lowest values.
This parameter requires a number greater than 0. If it is greater than 1, the selection is based on the numeric value. If it is between 0 and 1, the selection is based on a percentage.。
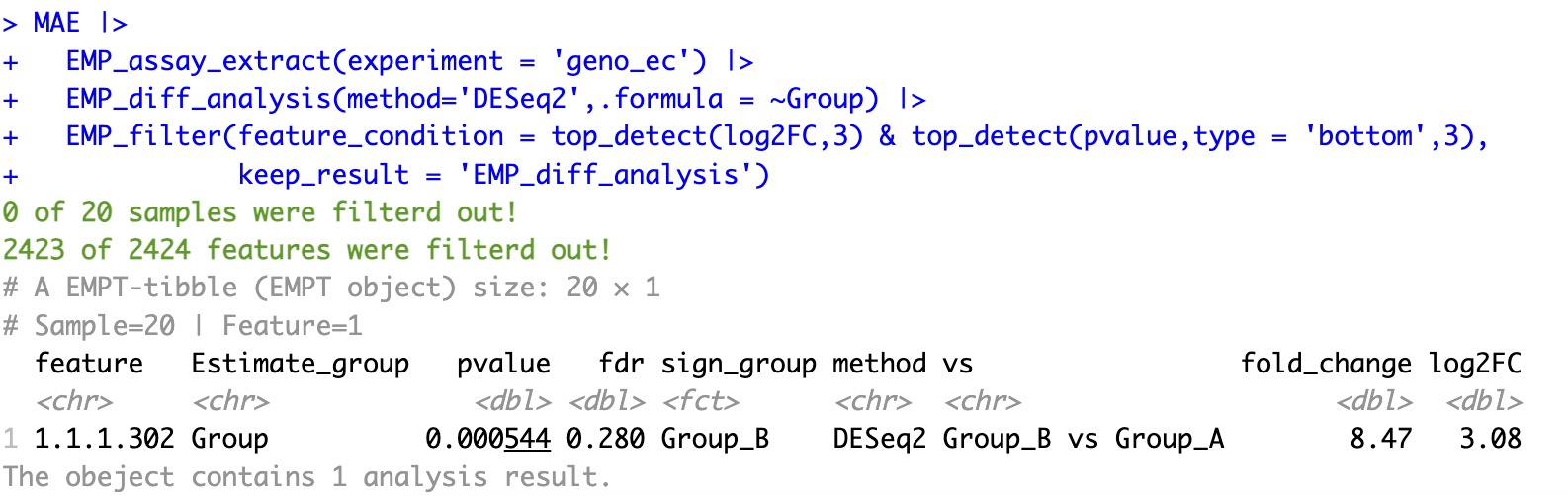
🏷️Example:
Select the intersection of the top three features with the highest log2FC and the top three features with the smallest p-value from the differential analysis.
MAE |>
EMP_assay_extract(experiment = 'geno_ec') |>
EMP_diff_analysis(method='DESeq2',.formula = ~Group) |>
EMP_filter(feature_condition = top_detect(log2FC,3) & top_detect(pvalue,type = 'bottom',3) ,
keep_result = 'EMP_diff_analysis')
9.4 Multi-group Differential Analysis with stat_test
This function enables rapid differential analysis comparisons among multiple groups for tabular data, automatically setting appropriate positional parameters to facilitate visualization with ggplot2.
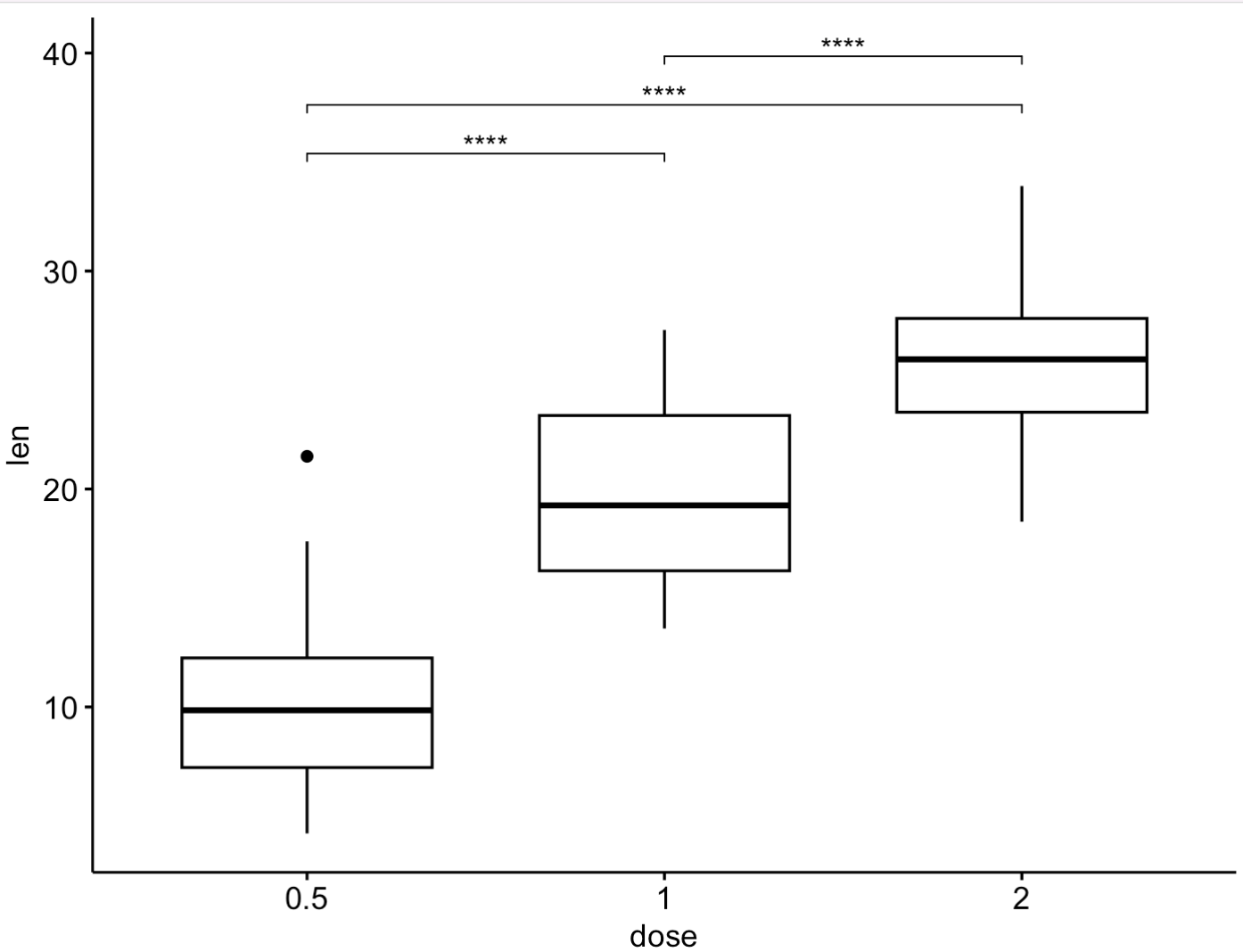
🏷️Example1: Standard comparison
data(ToothGrowth)
# Specifying grouping variable
stat_test(
data = ToothGrowth,
estimate_group = 'dose',
value = 'len',
method = "t.test"
)
# Using formula syntax
stat_test(
data = ToothGrowth,
formula = len ~ dose,
method = "t.test"
)
🏷️Example2: Subgroup comparison
data(ToothGrowth)
# Specifying grouping variables
stat_test(
data = ToothGrowth,
value = 'len',
estimate_group = "supp",
compare_group = "dose",
method = "t.test"
)
# Using formula syntax
stat_test(
data = ToothGrowth,
estimate_group = 'supp',
formula = len ~ dose,
method = "t.test"
)
🏷️Example3: Integration with ggplot2 visualizations
stat_result <- stat_test(
data = ToothGrowth,
formula = len ~ dose,
value = 'len',
method = "tukey.hsd",
)
# for ggpubr
if(require("ggpubr")){
ggboxplot(ToothGrowth, x = "dose", y = "len") +
stat_pvalue_manual(stat_result, label = "p.signif", tip.length = 0.01)
}